Link to original video by math et al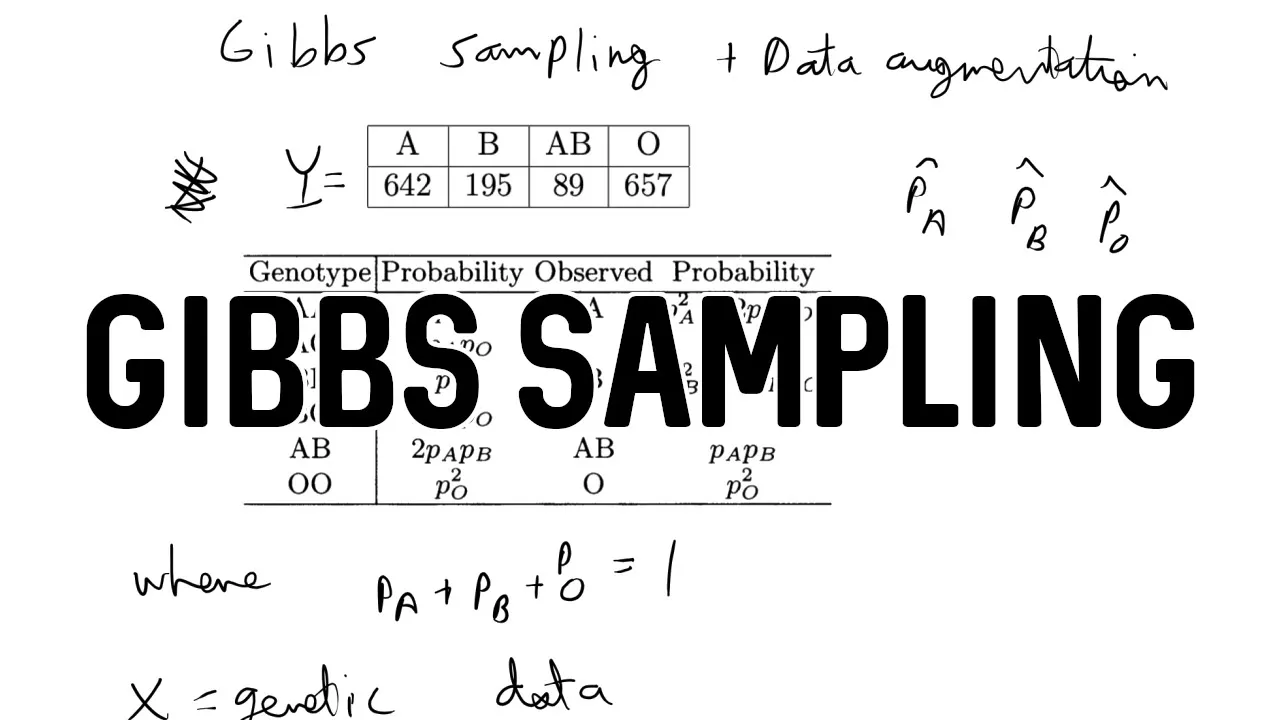
Gibbs Sampling and Data Augmentation w/ R Code | ABO Blood Typing Example

Summary of "Gibbs Sampling and Data Augmentation w/ R Code | ABO Blood Typing Example"
Short Summary:
- This video demonstrates Gibbs sampling, a Monte Carlo method, with data augmentation. The example used is estimating the genetic frequencies of blood types (A, B, O) from observed blood type data.
- The video explains how data augmentation addresses the issue of incomplete likelihood by introducing latent variables to represent missing genotype information.
- The process involves defining a posterior distribution, using a Dirichlet prior, and iteratively sampling from conditional distributions of latent variables and parameters until convergence.
- The video includes R code for implementing Gibbs sampling and visualizing the results.
Detailed Summary:
1. Introduction (0:00 - 1:20):
- The video introduces Gibbs sampling and data augmentation as tools for solving estimation problems.
- The specific example is estimating the genetic frequencies of blood types (A, B, O) from phenotypic data.
- The goal is to estimate the proportion of each allele (pA, pB, pO) using a Bayesian approach.
2. The Problem (1:20 - 2:10):
- The video explains the challenge of estimating genetic frequencies from observed blood types.
- The observed blood type (phenotype) is determined by the underlying genotype, which is not directly observed.
- This leads to an incomplete likelihood, making it difficult to estimate the parameters using traditional methods.
3. Data Augmentation (2:10 - 3:30):
- The video introduces data augmentation as a solution to the incomplete likelihood problem.
- Latent variables (Z1, Z2) are introduced to represent the missing genotype information for blood types A and B.
- By augmenting the data with these latent variables, a complete likelihood can be obtained.
4. Posterior Distribution (3:30 - 5:00):
- The video defines the posterior distribution of the parameters (pA, pB, pO) given the observed phenotypes and latent variables.
- A Dirichlet prior is chosen as the conjugate prior for the multinomial distribution of the genetic data.
- The posterior distribution is also Dirichlet distributed with updated parameters based on the observed data and latent variables.
5. Gibbs Sampling (5:00 - 7:30):
- The video explains the process of Gibbs sampling, an iterative Monte Carlo method for estimating the parameters.
- The process involves sampling from conditional distributions of the latent variables (Z1, Z2) and the parameters (pA, pB, pO).
- The estimates are updated iteratively until convergence, providing estimates for the genetic frequencies.
6. R Code Implementation (7:30 - 10:00):
- The video demonstrates the R code used to implement Gibbs sampling and visualize the results.
- The code includes functions for sampling from the Dirichlet distribution, initializing variables, and running the Gibbs sampler.
- The results show the estimated values for pA, pB, and pO, which are very close to the true values, indicating the success of the Gibbs sampling method.
7. Conclusion (10:00 - 10:30):
- The video concludes by summarizing the results and highlighting the effectiveness of Gibbs sampling with data augmentation in estimating genetic frequencies.
- The video emphasizes the importance of Bayesian statistics and Monte Carlo methods in addressing complex estimation problems.
Notable Quotes:
- "Data augmentation, as the name implies, basically means we're going to try to add to our sample data that we have. We're going to try to augment it to be able to form a complete likelihood." (2:10 - 2:30)
- "In Bayesian statistics when you don't know anything about alpha1, alpha2, and alpha3, which are the hyper-parameters, you can use what's called a noninformative or flat prior." (4:40 - 4:50)
- "And that's it for this video and thank you guys for watching." (10:10 - 10:20)